Referenced mature miRNA sequences
1,727 previously confirmed mature miRNA sequences of plants from miRBase release 12.0 were used as source sequences to identify potential miRNAs of an input sequence.
Input parameters
Sequence: a nucleotide sequence with less 3,000 bp
E-value : a parameter that describes the number of hits one can expect to see by chance when searching a database of a particular size (> 0)
Maximum number of alignments: a parameter that determines the maximum number of hit results (>0)
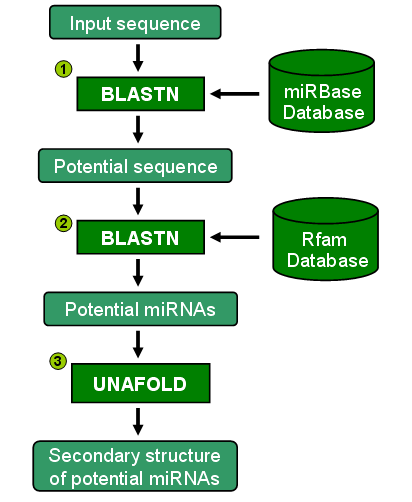
Prediction process
The prediction process as shown on the right hand side consists of three steps:
(1) The input sequence will be subjected to BLASTN program (version 2.2.18) to search for potential miRNAs against mature miRNA sequences referenced from miRBase (release 12.0). The user can specify an expected value (E-value) and a maximum number of alignments as a part of input parameters. The sequence that hits some miRNAs with the number of mismatches fewer than 3 will be identified as a potential sequence for the second step.
(2) The potential sequence will be subjected to BLASTN program to search for other types of RNA against RNA sequences referenced from Rfam database. The same parameters including an E-value and a maximum number of alignments are used as in previous step. The sequence that does not hit with other types of RNAs such as tRNAs or rRNAs will be identified as a potential miRNA coding sequence.
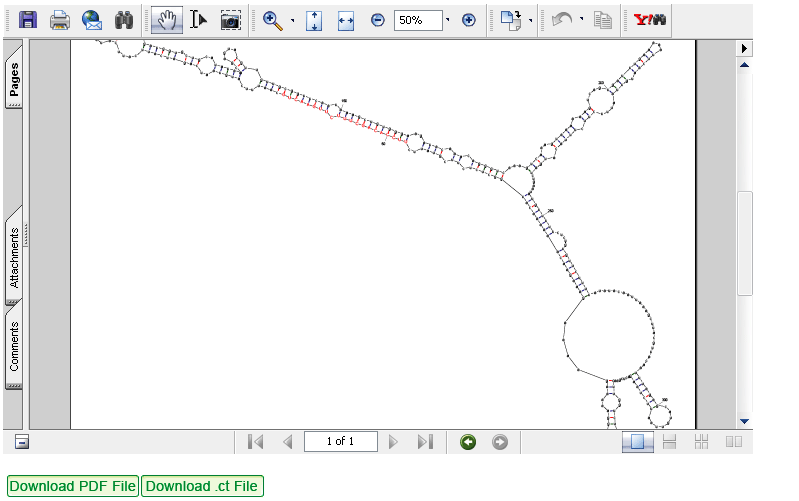
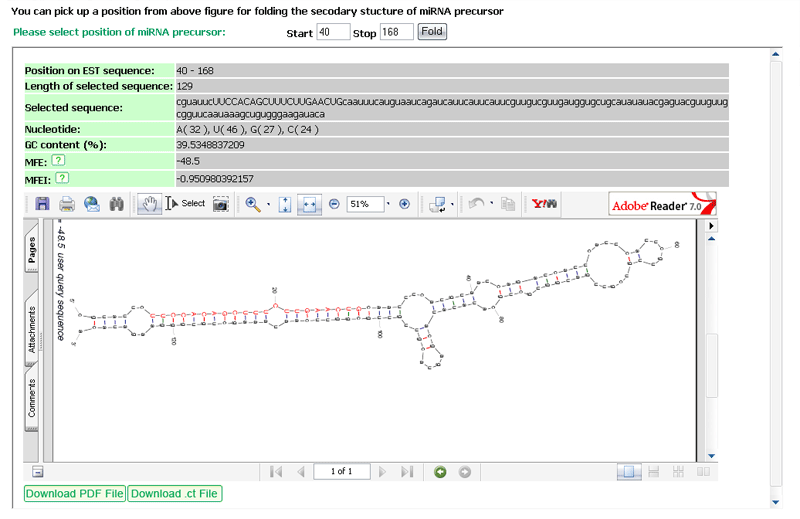
(3) The sequence from step 2 will be predicted for its secondary structures by UNAFold software (version 3.5) with default parameters.
Output format
Summary of input: includes input sequence length, E-value, maximum number of alignments, and data set (see example below)
| Summary of input |
| Length of input: 331 nucleotides |
| E-value: 0.1 |
| Maximum number of alignments: 50 |
| Data set: Plant mature miRNA sequences referenced from miRBase |
Summary of output: includes all mature miRNA sequences from miRBase that satisfy the setting conditions and found within the input sequence. These sequences will be represented by miRBase ID and AC, bit score, E-value, sequence alignment and linked to their homologs in the µPC. The input sequence with a specific mature miRNA positioned could be further predicted by UNAFold for its secondary structure (see below example).