Target Scanner |
| . |
Referenced target sequences EST sequences of 108 plants from PlantGDB (November 2008) were used as source sequences to scan for EST sequence potentially targeted by an input miRNA. Functions of predicted miRNA targets were assigned by BLASTX against UniProt protein database with E-value < e-20. |
|
||||||
Input parameters Sequence: a mature miRNA with 18 - 26 nucleotides (Only 'ATCGU' are valid sequence letters) Number of allowed mismatches: a parameter that specifies the number of mismatches (0-6) allowed between the input mature miRNA sequence and its complementary site on a potential targeted sequence Number of allowed GU pairs: a parameter that allows GU (Guanine:Uracil) pairs between an input sequence and its targets (0 - 4) Target data set: EST sequences of 108 plant species from PlantGDB database |
|||||||
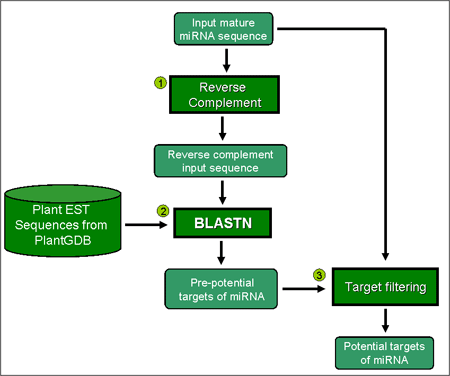
Prediction process The prediction process as shown on the right hand side consists of three steps: (1) The input mature miRNA will be reverse complement. (2) The reverse complement input sequence will be subjected to BLASTN program to search for pre-potential targets against the EST sequences in database. (3) The pre-potential miRNA targets will be filtered out by user specified parameters including numbers of allowed mismatches and GU pairs. |
|||||||
Output format Summary of input: includes input mature miRNA sequence, length, number of allowed mismatches, number of allowed GU pairs, and target data set (see example below)
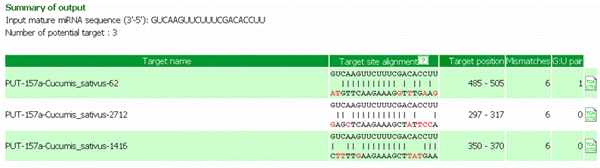
Summary of output: includes targeted EST accession, target site alignment and position, numbers of mismatches and G:U pairs and its functional annotation (see below example)
|
|||||||