miRNA tab
The miRNA tab allows users to explore the availability of miRNAs among plant species. Users may select miRNAs of interest by moving them from the left box to the right box (Figure 1).
It will return a matrix view of selected miRNAs and plants with cells colored purple, orange, or red respectively representing available miRNAs from our EST analysis, previous reports from miRBase, or both. Users may filter out plants without the selected miRNAs and group them according to their classification. Figure 2 shows a comparative matrix of miRNA 156, miRNA 159, miRNA 172, miRNA 444, miRNA 1062, miRNA 1076, miRNA 1082, miRNA 1091, miRNA 1094, miRNA 1095, and miRNA 1098 sorted by plant classification (e.g., monocots, eudicots) and filtered for only plants found with the query. The click on a purple or red cell will link to their detailed result in the µPC database.
Figure 1: Input of Comparative miRNAs
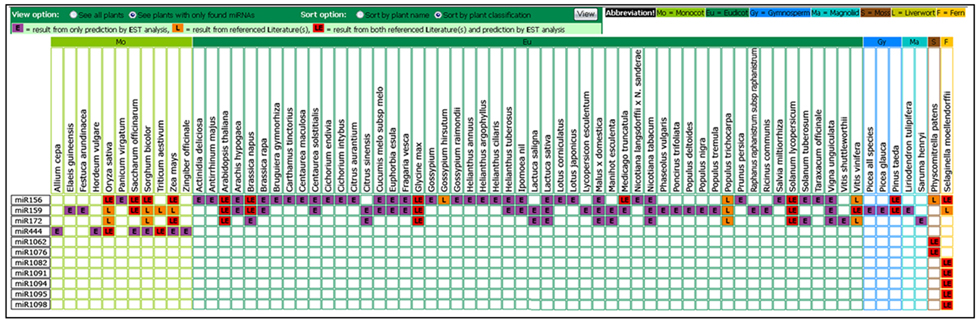
Figure 2: Output of Comparative miRNAs
sequence tab
The Sequence tab allows users to explore the alignment and evolution of either mature or precursor miRNA sequences from miRBase and our prediction results as well as optional user-supplied sequences. The maximum number of user-supplied sequences is 10 and the sequences need to be in FASTA format. Figure 3 shows the multiple sequence alignment by CLUSTALW in Jalview of precursor miRNA sequences from both miRBase and predicted EST sequences of miRNA397 family. Besides the alignment, users may explore the phylogenetic tree of the aligned sequences by using Jalview features under Calculate tab as shown in Figure 4.
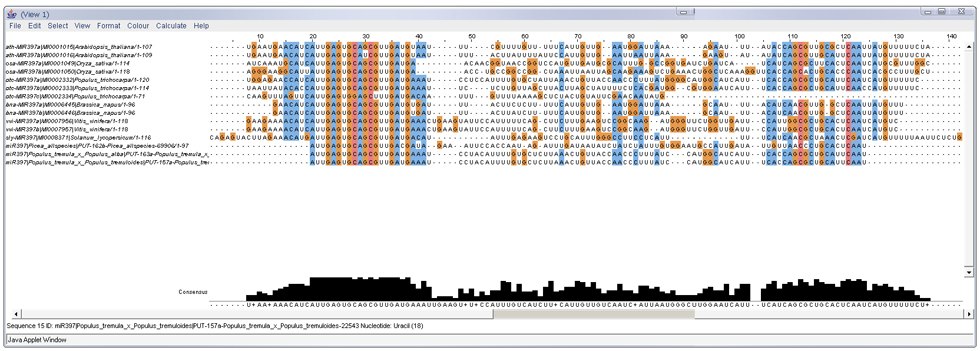
Figure 3: Alignment result of precursor miRNA sequences from both miRBase and predicted EST sequences of miRNA397 family
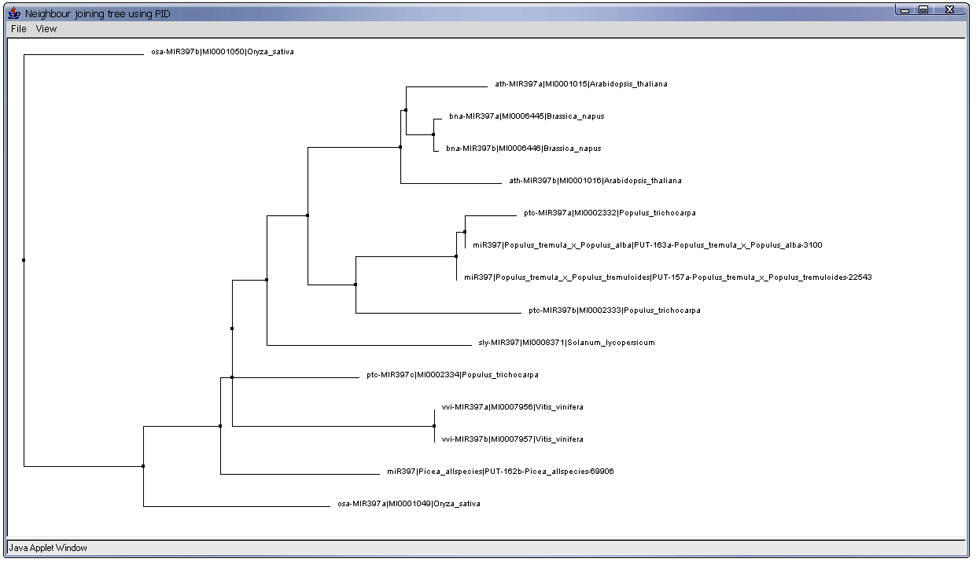 Figure 4: Building a phylogenetic tree from the aligned pre-miRNA sequences of miRNA397 family
Figure 4: Building a phylogenetic tree from the aligned pre-miRNA sequences of miRNA397 family