Under the miRNA tab, users could search for predicted miRNAs by miRNA family (e.g. miR156), plant species (e.g. Arabidopsis thaliana ), mature miRNA sequence (e.g. ugacagaagagagugagcac), and miRBase mature sequence accession (e.g. MIMAT0000166) (Figure 5).
The miRNA search result is varied according to a specific search option. It mainly includes a summary and a list of the results.
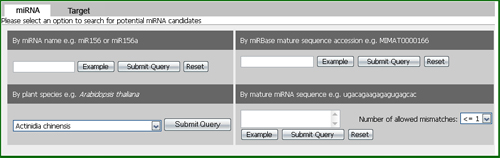
Figure 5: Search options
The summary in Figure 6A is resulted from a search by miRNA. The first line shows search option and its value. The second line shows the numbers of plant species, found with the searched miRNA, in total and by plant classification. The sorted by and filtered by options shows the current applied filters for the list of the results. The list of results (Figure 6B) includes the following fields:
|
Plant species: |
is the source species of the EST. |
|
miRNA: |
is the miRNA predicted to be encoded by the EST. |
|
EST name: |
is the PlantGDB accession number of the EST predicted as potential miRNA shown in the first field. |
|
Referenced source: |
lists the miRBase accession numbers that are sources of this predicted EST. The ‘*' following a miRBase accession number indicates its non-experimental source. The click on an accession number will link to its detailed information at miRBase website. |
|
Mature miRNA sequence: |
contains the mature miRNA sequence that corresponds to the referenced source miRNA. |
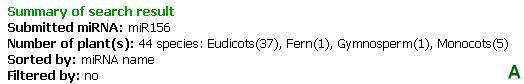
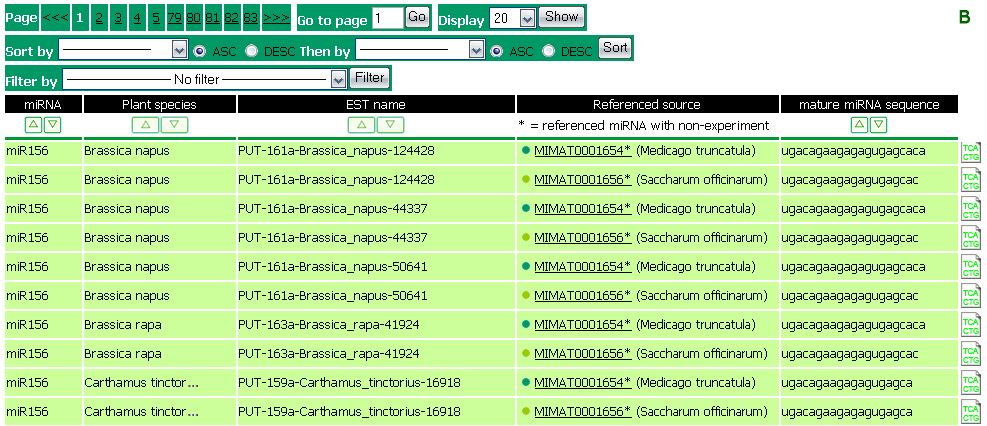
Figure 6: (A) Summary of search result and (B) the list of search results
The click on an icon ![]() will open a new window for the detailed prediction result including EST source sequence from PlantGDB, its secondary structure of pre-miRNA sequence, mature miRNA sequence, positions, number of mismatches, GC content, and MFEI (minimal folding free energy index) (Figure 7). The miRNA detail consists of the following fields:
will open a new window for the detailed prediction result including EST source sequence from PlantGDB, its secondary structure of pre-miRNA sequence, mature miRNA sequence, positions, number of mismatches, GC content, and MFEI (minimal folding free energy index) (Figure 7). The miRNA detail consists of the following fields:
Predicted miRNA name: |
is the name of miRNA that the EST predicted to encode. |
Plant species: |
is the plant species which is the source of the predicted EST. |
Plant taxonomy: |
is the plant classification of the sourced plant. EST name : is the EST accession number in PlantGDB. |
Length of mature miRNA: |
is the length of mature miRNA found in the EST |
Length of pre-miRNA: |
is the length of the precursor miRNA sequence of the EST. |
Location: |
is the strand of the EST found with the mature miRNA. |
Position on EST: |
is the position on the EST found with the mature miRNA. |
Mismatch: |
is the number of mismatches between the mature miRNA from miRBase and its sequence found on the EST. |
mature miRNA sequence (5' - 3'): |
is the mature miRNA sequence found on the EST. |
pre-miRNA sequence (5' - 3'): |
is the precursor miRNA sequence found on the EST. |
GC content of pre-miRNA sequence: |
is the % GC content over pre-miRNA sequence (number of G and C/number of all nucleotide)*100 |
MFEI: |
is the minimal folding free energy index calculated from the below formula MFEI = AMFE/(G+C)% where: AMFEI (Adjusted MFE) represents the MFE of 100 nucleotides, which was calculated by (MFE/length of RNA sequence)x100 and (G+C)% represents %GC content over pre-miRNA sequence. |
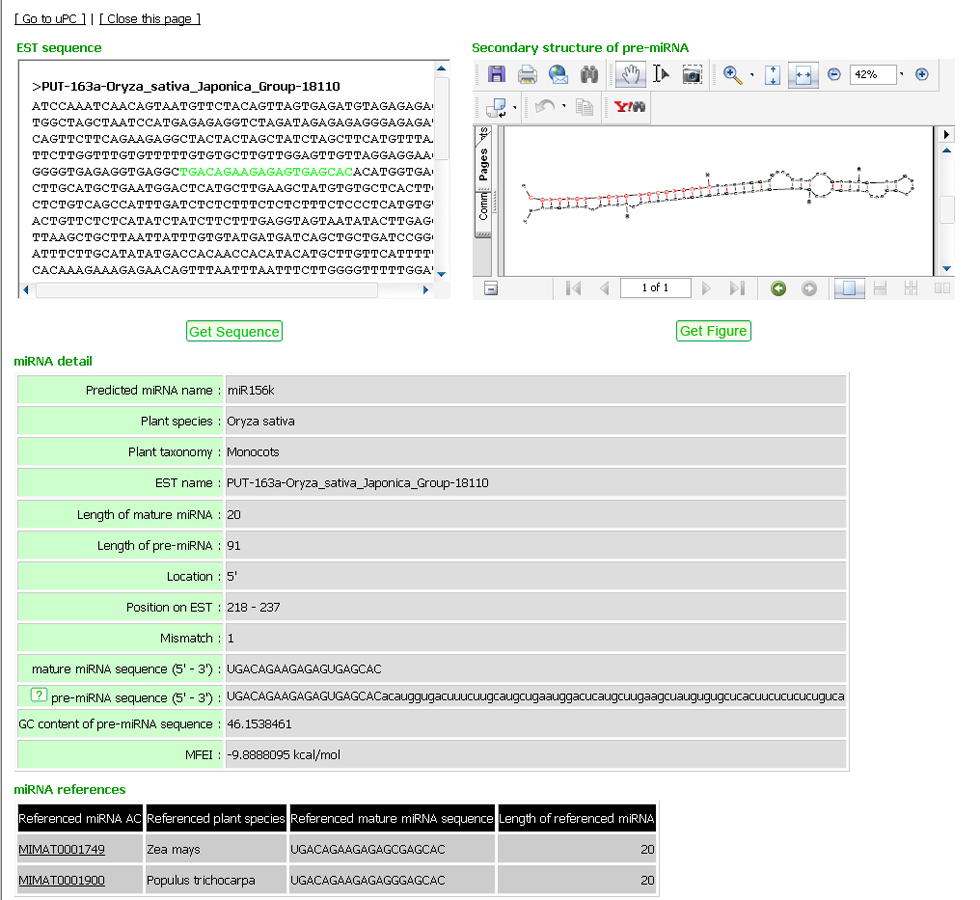
Figure 7: Detail of miRNA prediction result
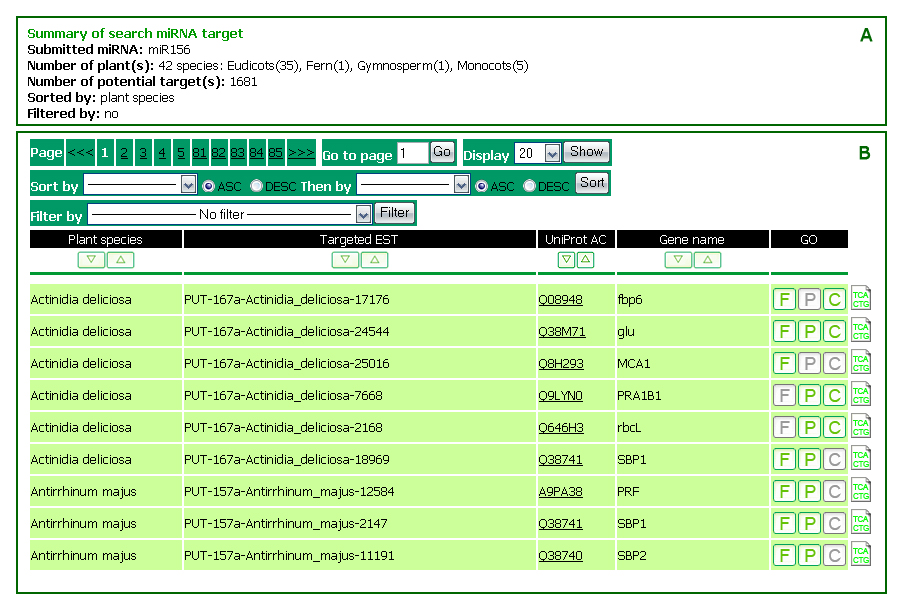
The Target tab allows users to search for ESTs potentially targeted by miRNA candidates through miRNA family, plant species, UniProt accession number (e.g. P93015), or GO term (e.g. rhythmic process). The target search result is varied according to a specific search option. It mainly includes a summary and a list of the results.
The summary in Figure 8A is resulted from a search by miRNA. The first line shows search option and its value. The second line shows the numbers of plant species having ESTs potentially targeted by the searched miRNA, in total and by plant classification. The sorted by and filtered by options shows the current applied filters for the list of the results.
The list of results (Figure 8B) includes the following fields:
Plant species: |
is the source species of the EST. |
Targeted EST: |
is the PlantGDB accession number of the EST potentially targeted by the search by miRNA name. |
UniProt AC: |
is the UniProt accession number of the EST resulted from BLASTX. The click on a UniProt AC will link to its detail information on the UniProt website. |
Gene name: |
is a common name of the UniProt AC. |
GO: |
is the Gene Ontology annotation for the predicted EST. F, P, and C respectively stand for Molecular Function, Biological Process, and Cellular Compartment as categorized by GO. A gray box of F, P, or C indicates its unavailable information. Users may see a GO annotation by moving mouse over a green box of F, P, or C. |
Figure 8 (A) Summary of target search by miRNA family and (B) the list of search results
The click on an icon ![]() will open a new window for the detailed prediction result including the source EST sequence from PlantGDB, the target alignment information, and the annotations.
will open a new window for the detailed prediction result including the source EST sequence from PlantGDB, the target alignment information, and the annotations.
The green sequence in the source EST sequence from PlantGDB represents the complementary (binding) site of the mature miRNA sequence (Figure 9A). The target site alignment (Figure 9B) shows the matches and mismatches of the mature miRNA sequence and its complementary site on the EST. The sequence with colors is the subsequence on the EST. The top sequence is the reverse complement sequence from the mature miRNA sequence in column three. The target annotation (Figure 9C) includes detailed information of BLASTX results, GO IDs, and GO terms annotated for the targeted EST.
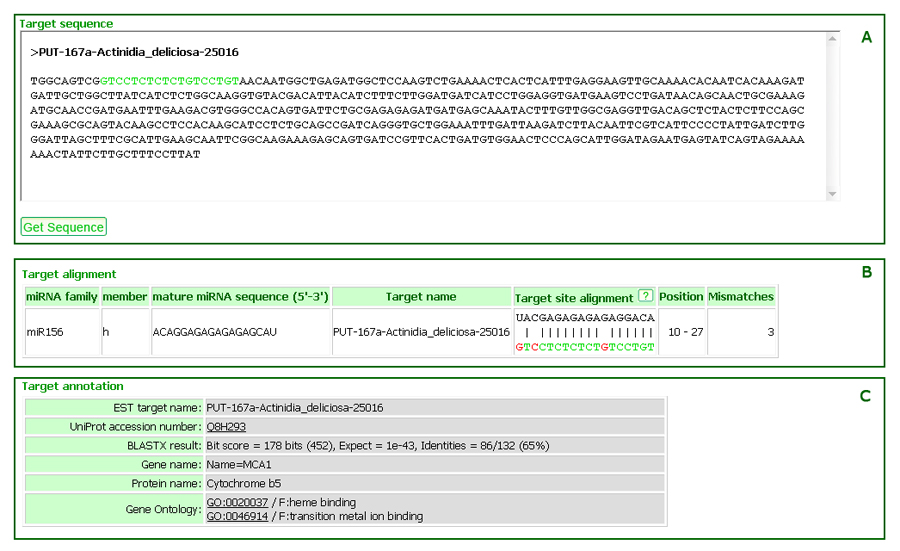
Figure 9: (A) Source sequence of targed EST with highlighted miRNA binding site, (B) target site alignment, and (C) target annotation.